Structural analysis
Structural Description
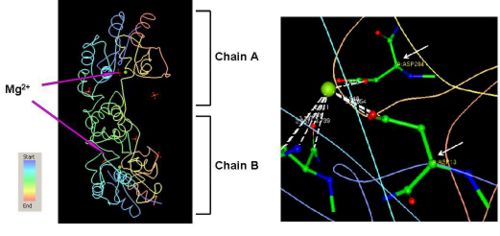
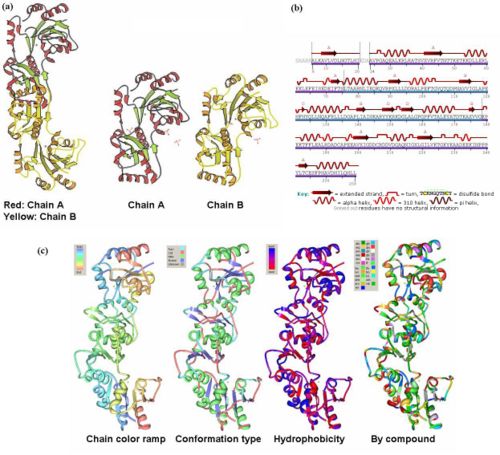
- The information on the PDB website suggested that 2HO4 is a hydrolase belonging to the haloacid dehalogenase (HAD) superfamily, consisting of 2 domains. It consists of 259 residues and has a molecular weight of 29 kDa. Moreover, the PDB structure viewers demonstrated that 2HO4 is a monomer but forms a dimer with itself to give Chain A and chain B (Fig 1a). The PDB workshop enabled us to examine the various properties of 2HO4 including its conformation type and hydrophobocity. Its secondary structure mainly consists of alpha helices and beta sheets with 44% helical (14 helices; 123 residues) 17% beta sheet (12 strands; 48 residues), and it consists of two domains (Fig 1b). The two chains bind near their hydrophobic regions to produce dimeric structures. The outer surfaces of the dimeric protein are mainly hydrophilic, while the ligand binding sites appear to be hydrophobic (Fig 1c). -
-
- - 2HO4 has two different ligands i.e a magnesium ion and three phosphate ions. The PDB structures of 2HO4 indicate that the Mg2+ ion binds between the coordinates Asp 13, Asn 15, Asp 204, and Asp 305 on each of the chains as shown in (Fig 2). -
-
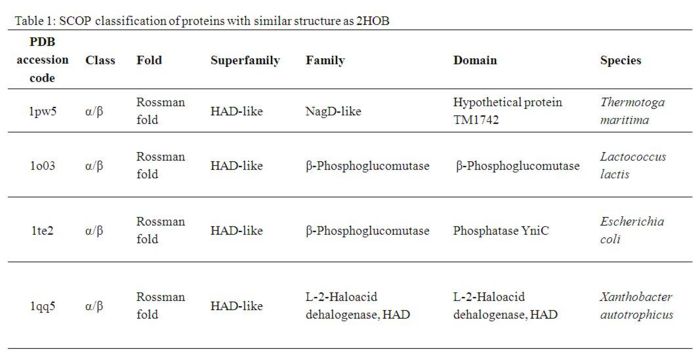
- - - == Structural Comparison == - - - A Dali search with a strict threshold of Z > 12 for 2HO4 found 10 matches. Of these 10 protein matches, we found that most of them were hydrolases belonging to various different families. We reduced our structural matches of 10 proteins to only the ones that had been previously annotated. These proteins had sequence identities within the range of 17% to 25% and they included a putative NagD-like protein of Thermotoga maritima, the β-phosphoglucomutase of Lactococcus lactis , the β-phosphoglucomutase of Escherichia coli, and the L-2-Haloacid dehalogenase of Xanthobacter autotrophicus. The results are summarized in Table 1. -
-
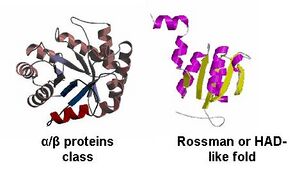
- - By looking at the architectural classification (using SCOP) of these proteins, it was noted that all of these proteins structures consisted of the same class, fold, and superfamily. The class was alpha and beta (α/β), which contains many parallel beta sheets (beta-alpha-beta units). Likewise, all the proteins had a Rossman fold (or HAD-like fold) consisting of three layers of α/β/α and parallel beta-sheets of 6 strands (Fig 3). And the superfamily they belonged to was HAD-like, which usually contains an insertion (sub)domain after strand 1. -
-
- - Literature seach indicated that members of the HAD superfamily included L-2-haloacid dehalogenases, sugar-phosphatases, and P-type ATPases, and all members shared three highly-conserved motifs. We did a multiple sequence analysis of H2O4 using the sequence homologs from BLAST as well as the structural homologs from DALI to show that it contains the three highly-conserved sequence motifs of the HAD superfamily. This confirmed that 2HO4 is a member of this superfamily (Fig 4a). The three motifs are: motif I: DXDX[T/V][L/N]; motif II: [S/T]XX; and motif III: K-[G/S][D/S]XXX[D/N]. These motifs lie next to each other and next the Mg2+ ion, together defining the active site of 2HO4 (Fig 4b) | (Rangarajan et al, 2006). -
-
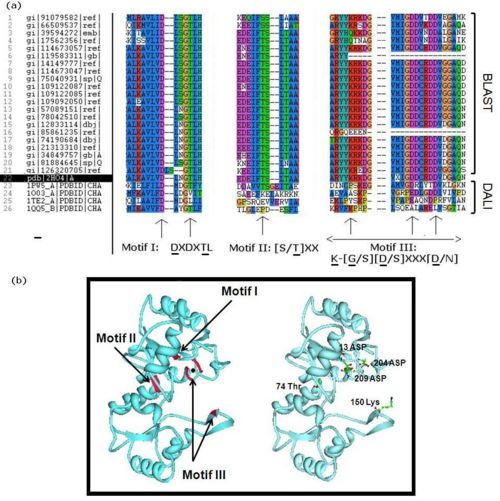
.
- - Furthermore, we investigated the alignment of the residues around the active site in space for any similarities. From the DALI results we used two well studied proteins: the β-Phosphoglucomutase of L. lactis (PDB code 1O03), and the L-2-Haloacid dehalogenase of X. autotrophicus (PDB code 1QQ5). We found that 2HO4 and β-Phosphoglucomutase of L. lactis had the three Asp residues from motifs I and III in common (Fig 5a), whereas 2HO4 and the L-2-Haloacid dehalogenase of X. autotrophicus had the Asp from motif I, the Ser/Thr from motif II, and the Lys from motif III in common (Fig 5b). Besides the architectural similarity between 2HO4 and β-Phosphoglucomutase of L. lactis (1O03), the L. lactis protein also consists of a Mg2+ ion bound to its catalytic site. It is possible that the two proteins share similar reaction mechanisms. -
-
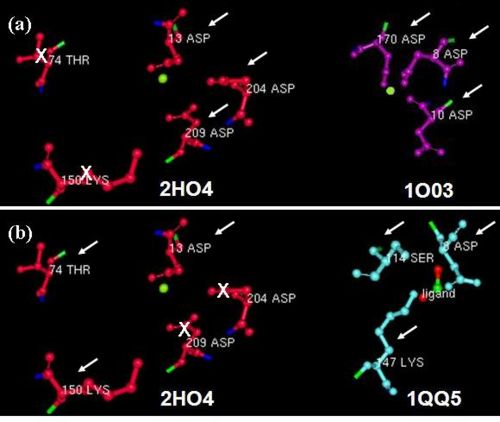
- - In order to determine which family of the HAD superfamily 2HO4 belonged to, we reviewed the literature. We found that HAD members with the conserved Asp residues and Mg2+ dependent reaction mechanisms could be P-type ATPases, which are involved in ATP hydrolysis. - - ---- - - | Back to report menu