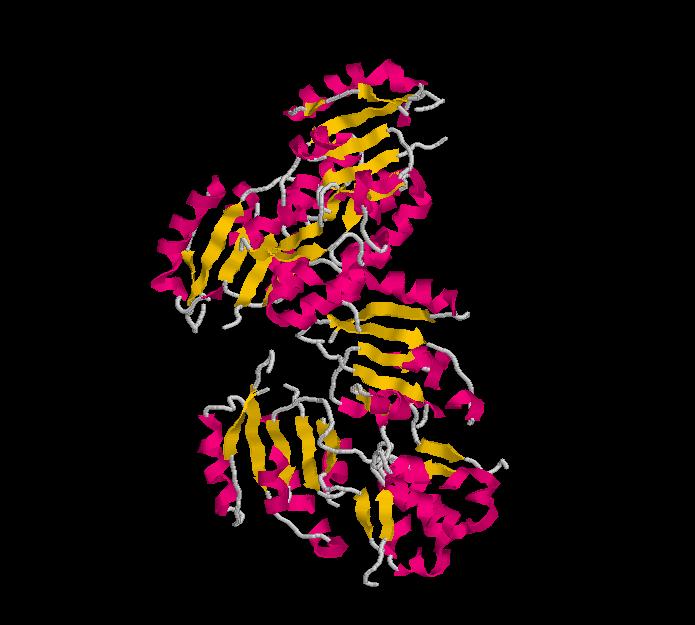
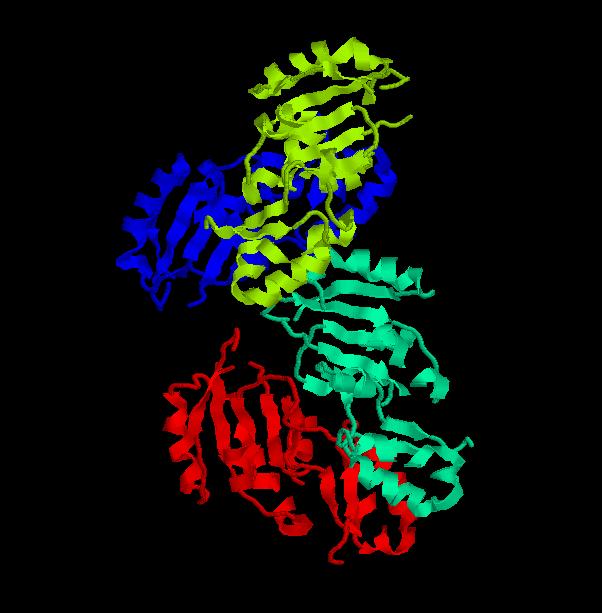
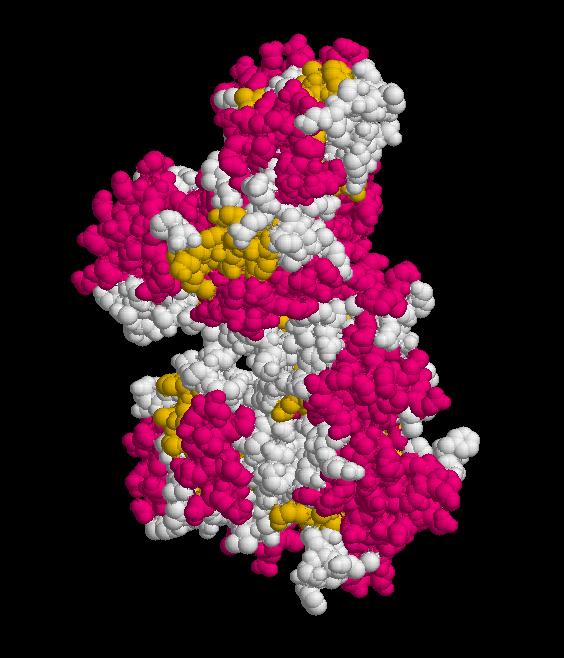
Structure.
Structure of Ssu72 RNA polymerase II CTD phosphatase homolog
PDB ID: 3fdf
Structure determined using XRD. The following images were obtained using RasMol.
Protein Structure Database Searching by DaliLite v. 3
The Dali server is a network service for comparing protein structures in 3D. You submit the coordinates of a query protein structure and Dali compares them against those in the Protein Data Bank (PDB).
These are the chains of different molecules that best matched to the Ssu72 RNA polymerase II CTD phosphatase homolog (it is important to notice that the first match corresponds to the protein that is being researched).
PDB ID Z rmsd lali nres %id PDB Description
3fdf-A 37.3 0.0 191 191 100 PDB MOLECULE: FR253;
3fmv-H 32.4 0.6 190 190 100 PDB MOLECULE: SERINE PHOSPHATASE OF RNA POLYMERASE II CTD;
1dg9-A 11.4 2.3 120 157 13 PDB MOLECULE: TYROSINE PHOSPHATASE;
1rxi-A 9.9 2.4 109 131 15 PDB MOLECULE: ARSENATE REDUCTASE;