Structure Protein: Difference between revisions
No edit summary |
No edit summary |
||
| Line 4: | Line 4: | ||
ligands: sulfate ion (SO4) and selenomethionine (MSE) | ligands: sulfate ion (SO4) and selenomethionine (MSE) | ||
10 sulfate ions were observed. Each ion differs in arrangement as well as orientation related with each chains. | 10 sulfate ions were observed. Each ion differs in arrangement as well as orientation related with each chains. | ||
Revision as of 03:04, 2 June 2007
2ifu has 4 chains (A, B, C and D) integrated together to form the whole molecular structure as shown below:
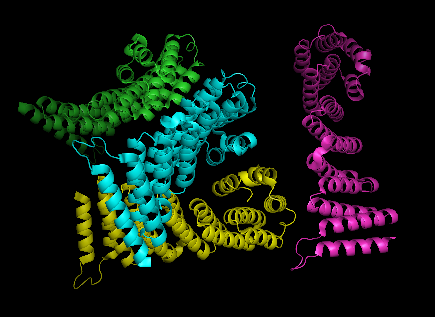
ligands: sulfate ion (SO4) and selenomethionine (MSE)
10 sulfate ions were observed. Each ion differs in arrangement as well as orientation related with each chains.
protein sequence (FASTA)
>gi|118138356|pdb|2IFU|A Chain A, Crystal Structure Of A Gamma-Snap From Danio Rerio AIAAQKISEAHEHIAKAEKYLKTSFXKWKPDYDSAASEYAKAAVAFKNAKQLEQAKDAYLQEAEAHANNR SLFHAAKAFEQAGXXLKDLQRXPEAVQYIEKASVXYVENGTPDTAAXALDRAGKLXEPLDLSKAVHLYQQ AAAVFENEERLRQAAELIGKASRLLVRQQKFDEAAASLQKEKSXYKEXENYPTCYKKCIAQVLVQLHRAD YVAAQKCVRESYSIPGFSGSEDCAALEDLLQAYDEQDEEQLLRVCRSPLVTYXDNDYAKLAISLKVPGGG GGKKKPSASASAQPQEEEDDEYAGGLC
Strutural comparisons
Based on Dali search, 5 most similar protein alignment was chosen: 1qqe-A (protein transport), 2fi7-A (protein binding), 1a17 (hydrolase), 2ak6-A, 1fch-A (signalling protein), 1haz4-A (transcription factor),
snap-gamma: multiple structure alignment
Snap-gamma: sequence alignment with 1QQE:A
Dark colors: 2ifu:A
Light colors: 1qqe:A
Protein families
has 2 domains: Pfam at Sanger