Structure Protein: Difference between revisions
No edit summary |
No edit summary |
||
| Line 87: | Line 87: | ||
http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi | http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi | ||
[[Image:ClustalX-alignment.JPG ]] | |||
Revision as of 06:29, 5 June 2007
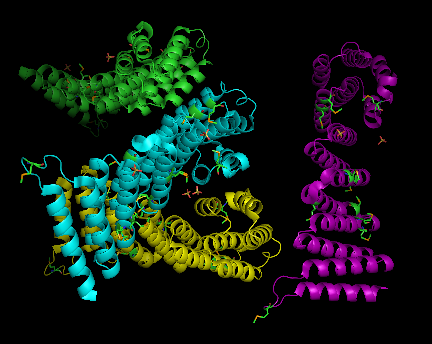
2ifu has 4 chains (A, B, C and D) integrated together to form the whole molecular structure as shown below:
(chain A= green, chain B= cyan, chain c= purple, and chain d= yellow)
Two molecular components were observed:
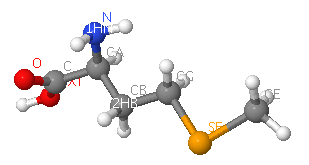
sulfate ion (SO4)and selenomethionine (MSE)
Ten sulfate ions were observed. Each ion differs in arrangement as well as orientation related with each chains.
protein sequence (FASTA)
>gi|118138356|pdb|2IFU|A Chain A, Crystal Structure Of A Gamma-Snap From Danio Rerio AIAAQKISEAHEHIAKAEKYLKTSFXKWKPDYDSAASEYAKAAVAFKNAKQLEQAKDAYLQEAEAHANNR SLFHAAKAFEQAGXXLKDLQRXPEAVQYIEKASVXYVENGTPDTAAXALDRAGKLXEPLDLSKAVHLYQQ AAAVFENEERLRQAAELIGKASRLLVRQQKFDEAAASLQKEKSXYKEXENYPTCYKKCIAQVLVQLHRAD YVAAQKCVRESYSIPGFSGSEDCAALEDLLQAYDEQDEEQLLRVCRSPLVTYXDNDYAKLAISLKVPGGG GGKKKPSASASAQPQEEEDDEYAGGLC
Strutural comparisons
Based on Dali search, 6 proteins with highest Z-value were chosen: 1qqe-A (protein transport), 2fi7-A (protein binding), 1a17 (hydrolase), 2ak6-A, 1fch-A (signalling protein), 1haz4-A (transcription factor),
snap-gamma: multiple structure alignment
Snap-gamma: sequence alignment with 1QQE:A
Dark colors: 2ifu:A
Light colors: 1qqe:A
SCOP
multiple alignment results, click here
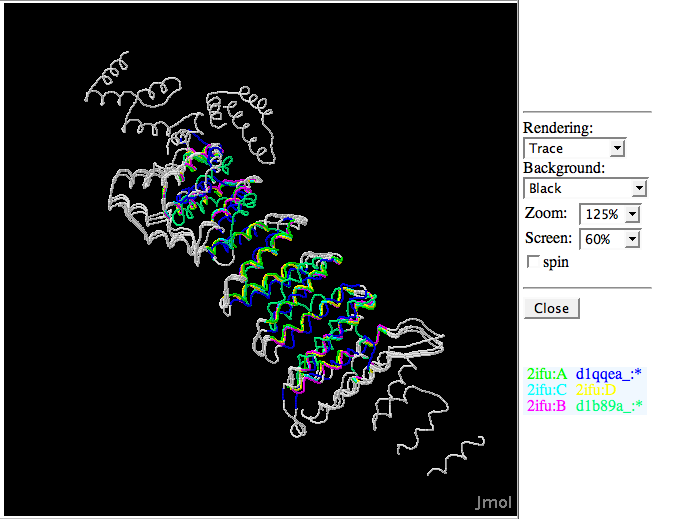
Superimposed structure between 2ifuA; 2ifuB; 2ifuC; 2ifuD; 1qqeA; and 1b89A
Protein families
has 2 domains: Pfam at Sanger
alignment with:
Pfam-B_7270 (7-66)
highly conserved residue:
Pfam-B_15198 (87-307)
highly conserved residue :residue 104, 109, 121, 168
scorecons result indicated a Diversity of position scores: 89.4% where 100%=high diversity
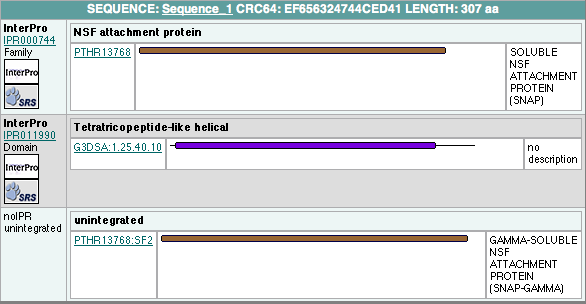
InterProScan Results
links
CRYSTAL STRUCTURE OF THE VESICULAR TRANSPORT PROTEIN SEC17 1QQE
CLATHRIN HEAVY CHAIN PROXIMAL LEG SEGMENT (BOVINE) 1B89
http://myhits.isb-sib.ch/cgi-bin/motif_scan
http://www.ncbi.nlm.nih.gov/Structure/vast/vastsrv.cgi?sdid=181604