View of the Binding Pocket: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
[[Image:surface view of ligand binding site.png|405px | [[Image:surface view of ligand binding site.png|405px]] | ||
[[Image:2NXF showing binding pocket4.png|405px|]] | |||
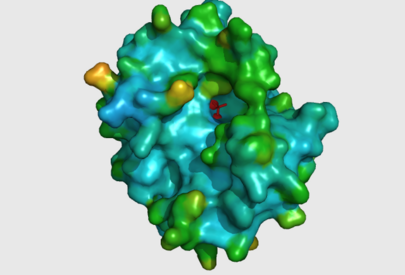
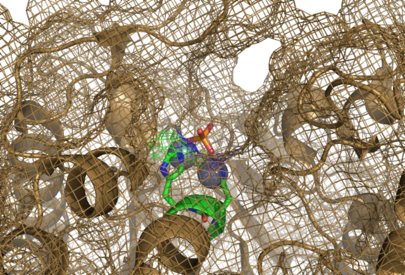
The large cavity of the 2nxf protein shown in a surface representation (left), showing its relative position with the rest of the protein. The ligands are shown here in red. The ribbon view, with mesh surface superimposed on top (right) is a close up of this same binding pocket, viewed from the right side of the protein. Analysis of the Dali results showed that the top 5 sequences contained three unchanging amino acid residues at positions 95 to 97 with a highly conserved acidic amino acid at position 98. The side-chains of conserved residues are shown here as coloured sticks. The two Zn atoms (spheres) appear to be indented into the surface of the protein with the phosphorous ion ligand sitting just above them on the surface of the protein. | The large cavity of the 2nxf protein shown in a surface representation (left), showing its relative position with the rest of the protein. The ligands are shown here in red. The ribbon view, with mesh surface superimposed on top (right) is a close up of this same binding pocket, viewed from the right side of the protein. Analysis of the Dali results showed that the top 5 sequences contained three unchanging amino acid residues at positions 95 to 97 with a highly conserved acidic amino acid at position 98. The side-chains of conserved residues are shown here as coloured sticks. The two Zn atoms (spheres) appear to be indented into the surface of the protein with the phosphorous ion ligand sitting just above them on the surface of the protein. | ||
Revision as of 15:28, 23 May 2008
The large cavity of the 2nxf protein shown in a surface representation (left), showing its relative position with the rest of the protein. The ligands are shown here in red. The ribbon view, with mesh surface superimposed on top (right) is a close up of this same binding pocket, viewed from the right side of the protein. Analysis of the Dali results showed that the top 5 sequences contained three unchanging amino acid residues at positions 95 to 97 with a highly conserved acidic amino acid at position 98. The side-chains of conserved residues are shown here as coloured sticks. The two Zn atoms (spheres) appear to be indented into the surface of the protein with the phosphorous ion ligand sitting just above them on the surface of the protein.