LOC56985 Structure Main: Difference between revisions
| Line 12: | Line 12: | ||
<gallery> | <gallery> | ||
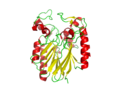
Image:2NXF ribbon structure .png|: Ribbon view of 2NXF showing the 4-layer sandwich structure described by the SCOP classification. | Image:2NXF ribbon structure .png|: [[Ribbon view of 2NXF]] showing the 4-layer sandwich structure described by the SCOP classification. | ||
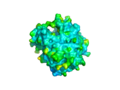
Image:2NXF surface by b factor.png|The surface view shows the large cavity, which is bridged at the top of the protein, many other cavities can also be seen. | Image:2NXF surface by b factor.png|The [[surface view]] shows the large cavity, which is bridged at the top of the protein, many other cavities can also be seen. | ||
Image:2NXF showing binding pocket4.png| View of the Binding Pocket with Zn and PO4 ligands | Image:2NXF showing binding pocket4.png| [[View of the Binding Pocket]] with Zn and PO4 ligands | ||
{..........} | {..........} | ||
</gallery> | </gallery> | ||
Revision as of 02:51, 21 May 2008
Structure Comparison
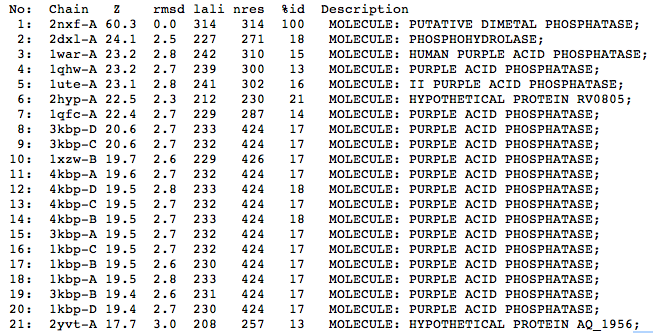
The Dali results indicate that structurally the closest relative to the protein is a phosphohydrolase (2DXL) from Enterbacter aerogenes. This protein shares similarity with 2NXF across 227 of 271 residues and shows a strong Z score of 24.1. The next closest matches were predominantly purple acid phosphatases from various mammalian species including humans, pigs and rats.
| Top 20 Structural Homologs Returned from the Dali Server |
|---|
Structure Analysis
The crystal structure of 2NXF shows that it binds 4Zn, PO4 and an ethanol molecule. The PO4 binds in the largest cleft in the protein at an active site that contains two Zn atoms. This site shows three identical residues across the top five protein matches by Dali. This includes the human homolog as well as the Enterbacter aerogenes protein. The sequence of residues is GNH, with a highly conserved acidic amino acid adjacent and down stream of this pattern. This same sequences were placed into the scorecons server to analyse how informative the alignment was; it returned a result of 99.2%, which rates this alignment as very diverse and informative.
- Ribbon view of 2NXF showing the 4-layer sandwich structure described by the SCOP classification.
The surface view shows the large cavity, which is bridged at the top of the protein, many other cavities can also be seen.
View of the Binding Pocket with Zn and PO4 ligands
Protein Architecture
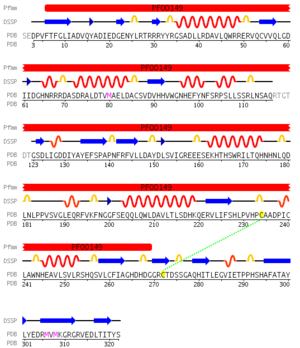
pfam analysis shows a common domain to all these proteins as being a calcineurin-like phosphoesterase (PF00149). SCOP classification of the protein shows it to be an Alpha and Beta (a + b) protein with a 4-layer sandwich fold of alpha/beta/beta/alpha of the Metallo-dependent phosphatases. it belongs to the serine/threonine phosphatase family of proteins. its CATH classification is 3.60.21.10.2.1.1.1.1.