Phytanoyl-CoA intro
Abstract | Introduction | Results | Discussion | Conclusion | Method | References
Phytanoyl-CoA dioxygenase domain containing 1 isoform a
Biochemistry, genetics and bioinformatics are three of the quickest growing fields in modern research. They have quickly established themselves over the last decade as better computer techniques have been made available. This increase in the quality of instrumentation has lead to an explosion of new genomic data to analyse, the majority of which is available online for public access. This report aims to elucidate, using bioinformatic databases, evolutionary, structural and functional information about the unannotated protein, phytanoyl-CoA dioxygenase domain containing 1 isoform a.
Target sequence -
>gi|134105315|pdb|2OPW|A Chain A, Crystal Structure Of Human Phytanoyl-Coa Dioxygenase Phyhd1 (Apo) MACLSPSQLQKFQQDGFLVLEGFLSAEECVAMQQRIGEIVAEMDVPLHCRTEFSTQEEEQLRAQGSTDYF LSSGDKIRFFFEKGVFDEKGNFLVPPEKSINKIGHALHAHDPVFKSITHSFKVQTLARSLGLQMPVVVQS MYIFKQPHFGGEVSPHQDASFLYTEPLGRVLGVWIAVEDATLENGCLWFIPGSHTSGVSRRMVRAPVGSA PGTSFLGSEPARDNSLFVPTPVQRGALVLIHGEVVHKSKQNLSDRSRQAYTFHLMEASGTTWSPENWLQP TAELPFPQLYT
The structure of phytanoyl-CoA dioxygenase domain containing 1 isoform a (PhyD) was determined using xray diffraction by Zhang et al. (2007) of the structural genomics consortium. Dioxygenases are oxidoreductases that oxidise a substrate by transferring both atoms of molecular oxygen. PhyD is structurally similar to other members of the phytanoyl-CoA dioxygenase family of proteins, which function in the peroxisomal pathway of alpha-oxidation.
Phytanoyl-CoA hydroxylase
The structure of one other member of the phytanoyl-CoA dioxygenase family, phytanoyl-CoA hydroxylase (PDB code 2a1x), has been determined. Phytanoyl-CoA hydroxylase contains an iron binding site, a 2-oxogluterate binding site and a N-terminus type-2 peroxisomal targeting sequence. This report compares the features of phytanoyl-CoA hydroxylase (PhyH) with PhyD to elucidate function.
Peroxisomal alpha-oxidation
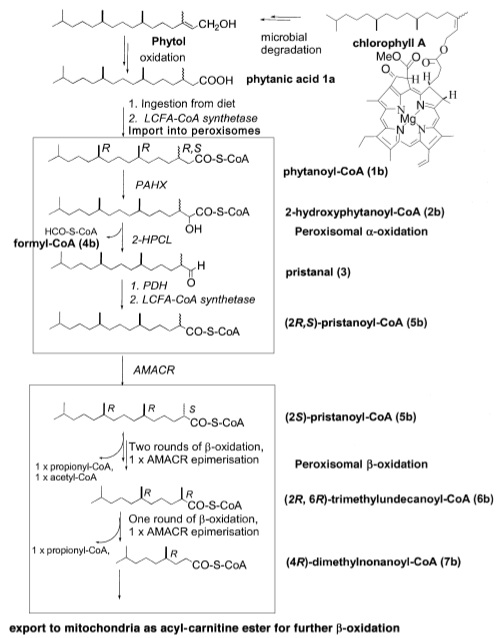
Phytanic acid is a long chain fatty acid constituent of the human diet. It is absorbed from a variety of foods, most notably the side chain of chlorophyll. The majority of long-chain fatty acids ingested in the diet are metabolised by beta-oxidation first in the peroxisome and then in the mitochondria. However, the methyl group on the third carbon of phytanic acid prevents beta-oxidation from occuring. Instead, the carboxyl group of phytanic acid is removed in alpha oxidation to allow for beta oxidation to take place.
Firstly, the phytanic acid undergoes the addition of co-enzyme A to the carboxyl group by long-chain fatty acid-CoA (LCFA-CoA) synthetase before being imported into the peroxisome. Once in the peroxisome the newly synthesised phytanoyl-CoA is metabolised by a phytanoyl-CoA dioxygenase to 2-hydroxyphytanoyl-CoA. Next, the alpha carbon is removed by 2-hydroxyphytanoyl-CoA lysase to form pristanal. Pristanal undergoes a further two reaction catalysed by pristanal dehydrogenase and LCFA-CoA synthase to form pristanoyl-CoA, a product able to enter the beta-oxidation pathway.
Refsum's disease
The result of defective alpha-oxidation is a build up of phytanic acid which manifests as Refsum's disease (RD). Mutations in phytanoyl-CoA hydroxylase have been shown to cause RD, charecterised by retinitis pigmentosa, anosmia, sensory neuropathy and phytanic acidaemia in adulthood. The symptoms of RD arise due to the build up of phytanoyl-CoA, which is inhibitory to some mitochondrial biochemical pathways.
Pyruvate dehydrogenase catalyses the production of acetyl-CoA from pyruvate for entry into the TCA cycle. The TCA cycle produces FADH2 and NADH as substrates for the electron transport chain. Acetyl-CoA is produced from pyruvate in a reaction catalysed by the pyruvate dehydrogenase complex. The pyruvate dehydrogenase complex is composed of the three enzymes (E1, E2 and E3) that produce acetyl-CoA in three defined, sequential reactions. E1 is inhibited by high concentrations of phytanoyl-CoA reducing the input of substrate into the TCA cycle (Bunik et al., 2006).
The 2-oxoglutarate dehydrogenase complex is similar in structure to the pyruvate dehydrogenase complex and also contains three enzymatic subunits. E1 of the 2-oxoglutarate complex is also inhibited by phytanoyl-CoA (Bunik et al., 2006). This situation is benificial to a cell with normally functioning phytanoyl-CoA hydroxylase as 2-oxoglutarate is an essential reactant in the oxidation of phytanoyl-CoA.
The result of inhibition of 2-oxoglutarate- and pyruvate- dehydrogenase is a reduction in TCA cycle activity and therefore reduced ATP production.
Abstract | Introduction | Results | Discussion | Conclusion | Method | References