Phytanoyl-CoA results: Difference between revisions
| Line 9: | Line 9: | ||
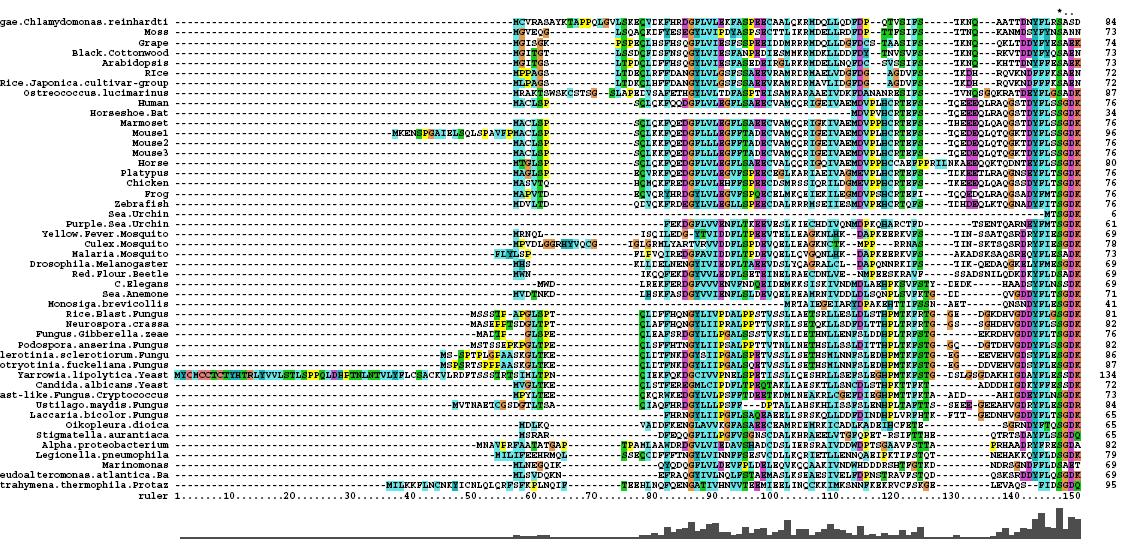
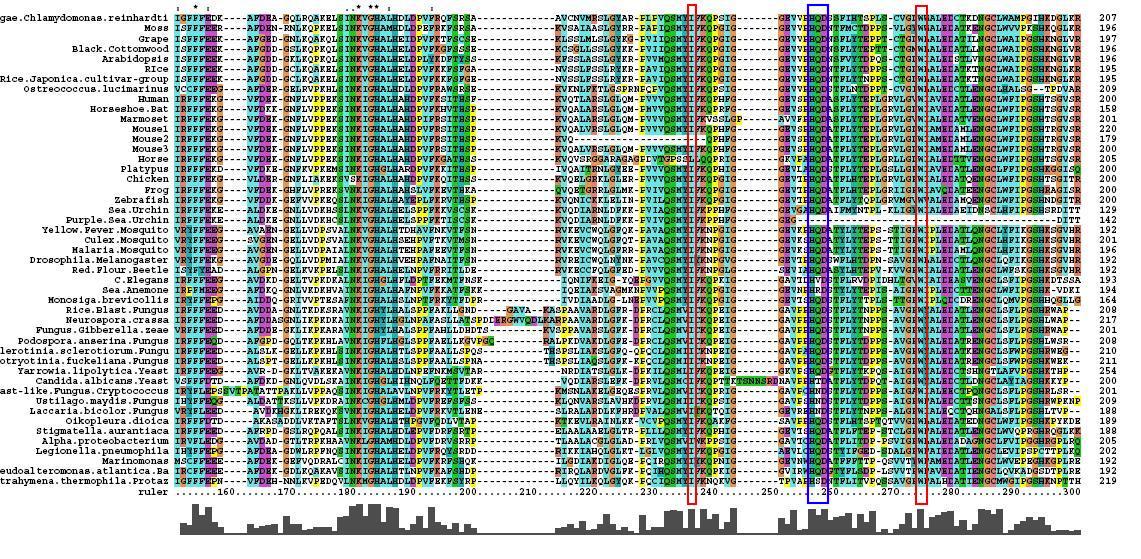
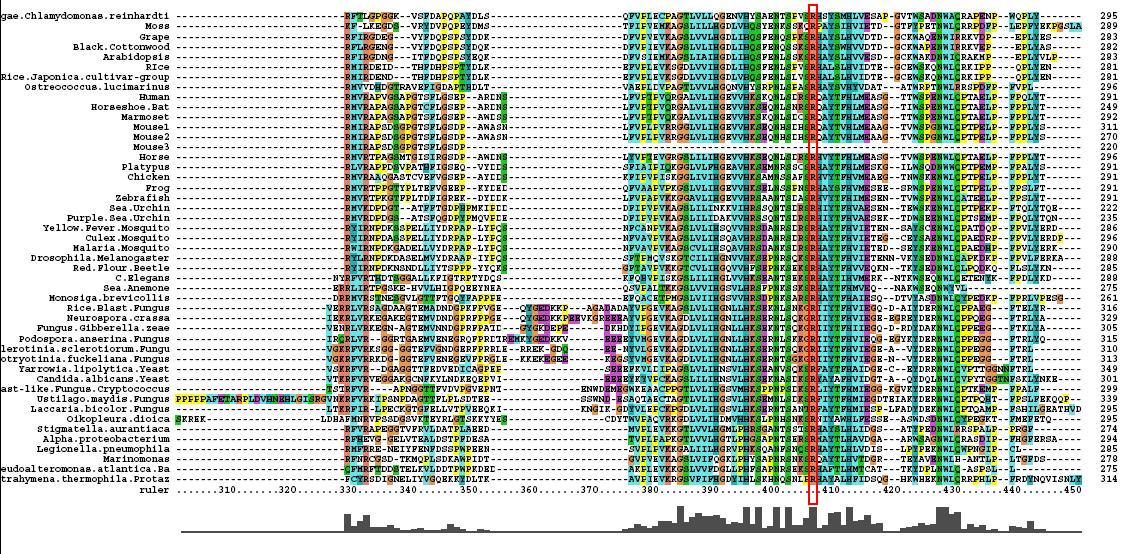
[[Image:Msa3.jpg|center|framed|'''Figure 1.3'''<Br> Multiple Sequence Alignment obtained from ClustalX]]<Br> | [[Image:Msa3.jpg|center|framed|'''Figure 1.3'''<Br> Multiple Sequence Alignment obtained from ClustalX]]<Br> | ||
[[Phytanoyl-CoA abstract | Abstract]] | [[Phytanoyl-CoA intro| Introduction]] | [[Phytanoyl-CoA results| Results]] | [[Phytanoyl-CoA discussion| Discussion]] | [[Phytanoyl-CoA conclusion| Conclusion]] | [[Phytanoyl-CoA method| Method]] | [[Phytanoyl-CoA references| References]] | |||
[[Phytanoyl-CoA dioxygenase domain containing 1 isoform a | Return to main page...]] | [[Phytanoyl-CoA dioxygenase domain containing 1 isoform a | Return to main page...]] | ||
Revision as of 23:55, 6 June 2008
Sequence Analysis
After conducting a BLAST search of the non-redundant databases, 46 analygous sequences were selected for comparison. A multiple sequence alignment was then performed on the sequences. The returned alignment was of an excellent quality with several section of high conservation being identified. The gene appears to be fairly well conserved across all species although the best similarity occurs with other mammalian species. This idicates that the protein is most likely to still have its functionality in most of these species which is remarkable given the broad range of both prokaryotes and eukaryotes investigated. In total there 5 residues conserved across all sequences but several more section of very high similarity. These include the iron binding sites (shown in red) and the 2OG binding site (shown in blue) as highlighted in Figure 1.
Abstract | Introduction | Results | Discussion | Conclusion | Method | References